Polymerase Chain Reaction (PCR)
Polymerase chain Reaction Protocol:
The Basic aim of a PCR (Polymerase Chain Reaction) is to make a huge number of copies of a gene. Molecular markers specific for Late blight, PVY and PCN are routinely used/tested for marker assisted molecular breeding.
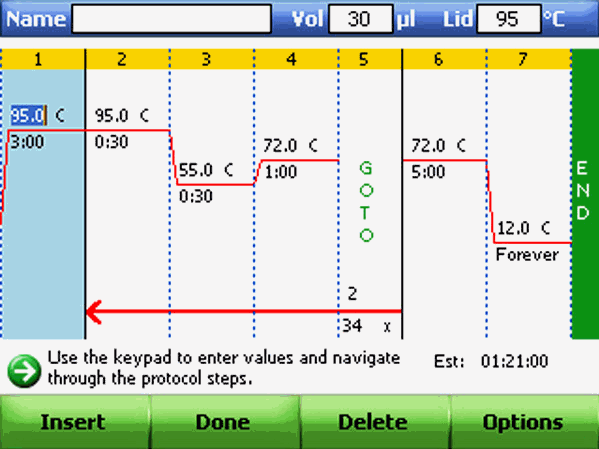
Cycle Conditions:
When first standardizing a PCR, it is suggested/recommended to do a temperature gradient.
Following guidelines can be useful while designing PCR protocol. This is done in an automated thermocycler which heats and cools the microtubes having PCR reaction mixture.
|
PCR Conditions |
Guidelines |
|
Denaturation |
Temp: 95° C. Time: 5 min on initial cycle; 30 seconds to 1 min on rest. (During the denaturation, the double strand nucleic acid gene template melts open to single stranded DNA). |
|
Annealing |
Temp: 5° C below Tm of primers; no lower than 40° C. Time: 30-45 seconds. (This is the step where a user may select a gradient temperature for specific amplification). |
|
Extension |
Temp: 72° C. Time: ~1 min/kb of expected product; 5-10 min on last cycle. |
|
Number of Cycles |
~30 cycles |
If, after trying both temperature and magnesium gradients (and checked all PCR reagents) PCR still does not work. Following additives may be used in the PCR master mix
Creating a New PCR Protocol or Editing an Existing Protocol in C-1000 thermocycler BioRad:
To create or edit a protocol: Select New Protocol from the main menu or press EDIT and select the protocol to be modified. A protocol will open in graphical view.


Inserting a Protocol Step:
Insert a protocol step if a new temperature, GOTO, or gradient step is needed. Follow these instructions to insert a step to the right of a pre existing protocol step:
1) Select a step to the left of where the new step is going to be inserted. Then click Insert.
2) Choose the type of step to insert. Select Temp to insert a temperature step, GOTO to insert a GOTO step, or Gradient to insert a gradient step.
3) Select and edit the parameters in the new step.
Adding or Removing a Temperature Gradient : To add a temperature gradient to a temperature step: 1. Select a step in the protocol and click Options. The Step Options window opens as follows.
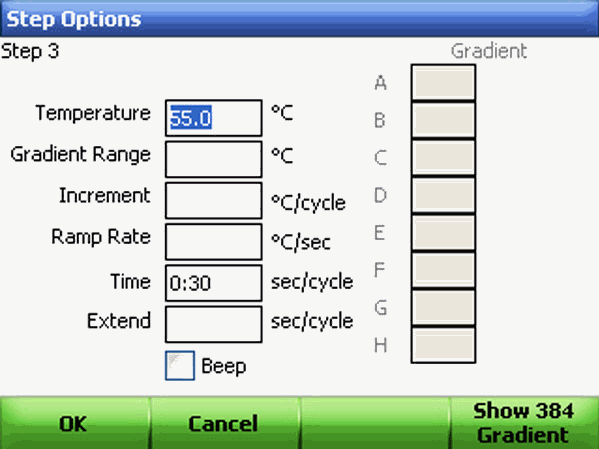
Using the alphanumeric keys, enter a value in the Gradient Range field. A gradient range must be between 1 and 24°C. The highest temperature in the gradient is the sum of the lower temperature and the gradient range. Once the gradient range is entered, the Step Options screen displays the gradient distributed across the rows of wells in the block. The gradient is displayed across a 96-well block from the back (row A, upper temperature) to the front (row H, lower temperature). Click OK to continue.
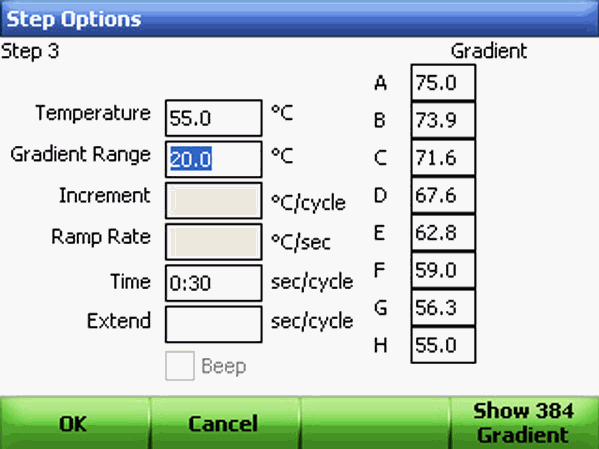
Review the gradient selection and once a step has a gradient, the upper and lower temperatures can be edited in the graphical or textual view without opening the Step Options screen.
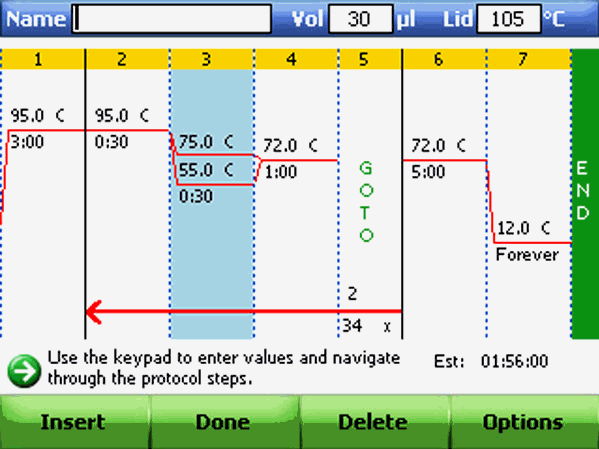
Adding or Removing a Temperature Increment
An increment raises or lowers the target temperature for a step with each cycle. To add or remove a temperature increment: Follow steps of "Adding or Removing a Temperature Gradient" but change the value in the Increment field instead. Review the increment in the textual view of a protocol.
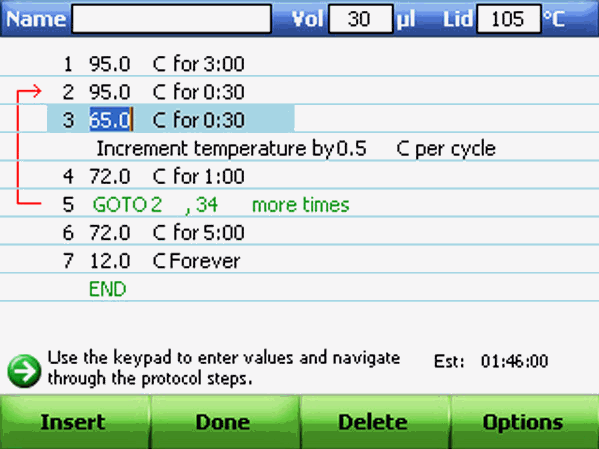
To remove an increment, select a step in the protocol and then select Options. The Step Options window opens. Delete the increment temperature by pressing BACK. 4. Select OK to confirm.
Changing Parameters in a GOTO Step : The GOTO step instructs the thermal cycler to repeat a set of steps in a loop. This step creates a cycle in the PCR experiment. To change parameters in a GOTO step, select a GOTO step in the graphical or textual view of the protocol. Press the arrow keys to select the number in the GOTO step. View the changes in the GOTO parameters in textual or graphical view.